 |
| Fig. 1 The Real RNA |
I recently decided to use it as the main format for translating DNA segments into mRNA, codons, and amino acids.
In all types of cells, the ribosome is composed of two subunits: the large (50S) subunit and the small (30S) subunit (S, for svedberg unit, is a measure of sedimentation velocity and, therefore, mass). Each subunit exists separately in the cytoplasm, but the two join together on the mRNA molecule. The ribosomal subunits contain proteins and specialized RNA molecules—specifically, ribosomal RNA (rRNA) and transfer RNA (tRNA). The tRNA molecules are adaptor molecules—they have one end that can read the triplet code in the mRNA through complementary base-pairing, and another end that attaches to a specific amino acid (Chapeville et al., 1962; Grunberger et al., 1969). The idea that tRNA was an adaptor molecule was first proposed by Francis Crick, co-discoverer of DNA structure, who did much of the key work in deciphering the genetic code (Crick, 1958).
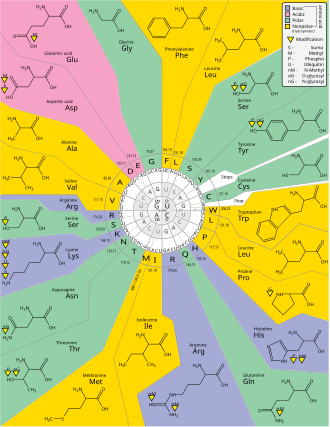 |
| Fig. 2 Genetic Code Perspective |
Within the ribosome, the mRNA and aminoacyl-tRNA complexes are held together closely, which facilitates base-pairing. The rRNA catalyzes the attachment of each new amino acid to the growing chain."
(Nature, Translation: DNA to mRNA to Protein). I will point out one atomic aspect of the process later on.
I has to do with thymine being converted to uracil during the process:
"Like DNA, RNA is a linear polymer made of four different types of nucleotide subunits linked together by phosphodiester bonds (Figure 6-4). It differs from DNA chemically in two respects: (1) the nucleotides in RNA are ribonucleotides—that is, they contain the sugar ribose (hence the name ribonucleic acid) rather than deoxyribose; (2) although, like DNA, RNA contains the bases adenine (A), guanine (G), and cytosine (C), it contains the base uracil (U) instead of the thymine (T) in DNA. Since U, like T, can base-pair by hydrogen-bonding with A (Figure 6-5), the complementary base-pairing properties described for DNA in Chapters 4 and 5 apply also to RNA (in RNA, G pairs with C, and A pairs with U). It is not uncommon, however, to find other types of base pairs in RNA: for example, G pairing with U occasionally."
(Molecular Biology of the Cell. 4th edition). As I have written previously, the GenBank nomenclature does not distinguish between thymine ("T") and uracil ("U") (It's In The GenBank - 4).
I added a feature to the software I engineered, which is the addition of the atomic nomenclature of the codons.
Anyway, the next step after converting the 'five-prime' (5`) and 'three-prime' (3`) segments (strands) into mRNA format codons ("U" for uracil, see Fig. 1 above).
Those codons can then be used to determine the amino acid using the Genetic Code (see Fig. 2).
As I said, I added the atomic nomenclature).
Here is an example from the software's analysis of chromosome 1:
output-files/chr1_fna.html
processing: >CM000663.2 Homo sapiens chromosome 1:
GRCh38 reference primary assembly
strand_53 ok, strand_35 ok, mRNAstrand ok
AGA,UGA,AGG,CUG,UCA,AGU,UAG,AGC,CUG,UCC,
AGC,AGA,CUG,AGA,AGA,UGA,CUG,CUC,CUA,CUG,
UUG,AGG,CUG,UAA,
AUG,CAU,CUU,CAC,UCC,CUC,AGA,AGG,AGG,CUC,
AUG,CUG,AGG,AGG,AGA,AGU,AGU,CGU,AGA,AAU,
AUG,CAU,UGA,CUU,AGG,CUC,UCU,CAU,AGG,AGG,
CUU,CUC,CUG,CAC,CUU,CUU,AGA,AGC,CAU,CUG,
CUA,CUG,CUA,UAA,
AUG,CUG,AGG,AGG,AGA,AGU,AGU,CGU,AGA,AAU,
AUG,CAU,CUU,AGG,CUC,UCU,CAU,AGG,AGG,CUU,
AUG,CUG,UUG,UCA,UCU,UCC,UGA,UUG,CUC,
AUG,CUG,UUG,UCA,UCU,UCC,UUG,CUC,
RLSS
SLSSRLRR
LLLLLRL
MLRRRSSRRNMH
LRLSHRRLLLHLLRSHLLLL
MLLSSS
LL
GCA,CCA,GCU,GUC,GAC,GGG,GAG,AAG,GGA,UGC,
CCU,GAU,ACA,GUG,AAA,AUU,CCG,AUC,UUC,GCG,
(Excerpts from chromosome 1). The molecular descriptions in the "molecule content" section are in red letters and digits ; the abbreviations mean: (MET="Methionine", HIS="Histidine", SER="Serine", ARG="Arginine", ASN="Asparagine", LEU="Leucine", and TYR="Tyrosine").
In the "valid codons" section the start and stop codons are in bold letters.
Notice that the software I engineered separates the mutant codons and places them into the "codon mutations" section of the analysis (only one copy of each mutant is displayed).
Proton tunneling has been a suspect in mutant phenomena for some time, so the "codon mutations" section is not useless (The Doll As Metaphor - 4).
More discussion of that and other issues will be forthcoming in future posts in this Dredd Blog series.
The next post in this series is here, the previous post in this series is here.
Translation and transcription video:
No comments:
Post a Comment