 |
Fig. 1 Nucleotides |
I. In General
The "ABC's" ("ACGT") of our genes are depicted in Fig. 1.
DNA genes are made of these molecules, excluding Uracil, which is in RNA.
The vast GenBank stores files of 'sequences' of these nucleotides (GenBank FTP download site).
The GenBank "Readme" file explains technical aspects of the site (Readme.genbank link to text file).
The process which a 'submitter' goes through to upload data into the database is explained here.
II. Accuracy Tests
Today's post contains a list of HTML tables in several appendices which show the results of Dredd Blog tests done on many of the files in the GenBank (Appendix A, B, C, D, E, F, G, H, I, J, and K).
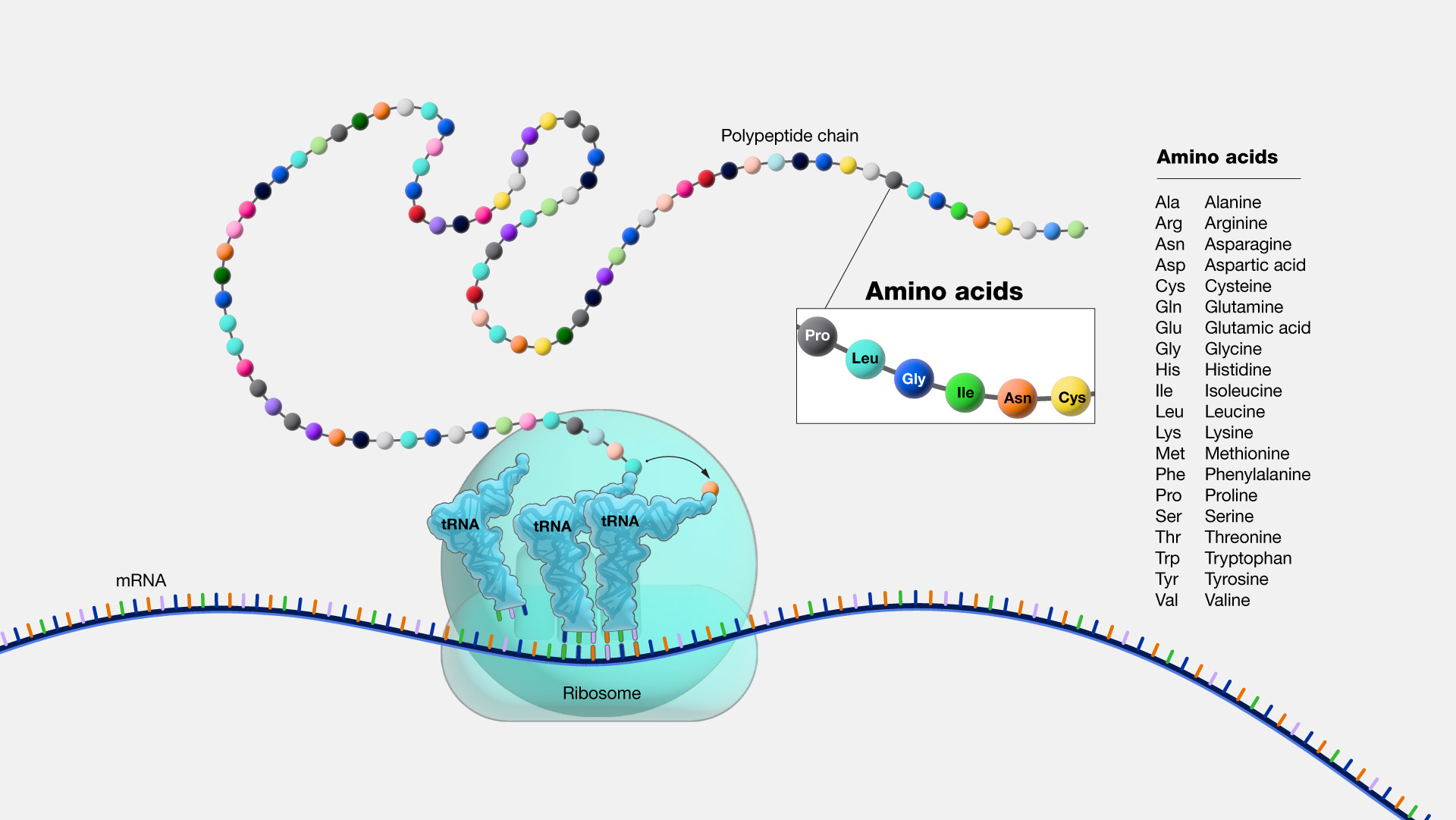 |
| Fig. 2 Amino Acids |
While the GenBank files are generally accurate, some of them have errors in the CDS->Translation data.
That is where, in the GBFF files, gene locations like "CDS 15..345" (which refers to nucleotide locations "fifteen through three-hundred and forty-five").
Codons within that range are 'translated' into an amino-acid sequence.
Amino acids are 'generated' by ribosomes based on the three nucleotide codons within the 15.345 range.
The "translation" results in a list of amino acids that are indicated by the codons in the 15..345 range of nucleotides (the 20 available amino acids are shown in Fig. 2).
In the CDS 15..345 example, that hypothetical list is 110 "codons" that are each made up of three of the ACGT molecules shown in Fig. 1.
The arithmetic is simple (345−15) ÷ 3 = 110 codons; so that CDS reference should equate to 110 amino acids (one amino acid per codon).
The "translation key" ("/translation=") in the GBFF files would refer back to the 110 letters (amino acids) each of which is derived from a three-letter codon by the sequencing machines and researcher's processing of the genetic material.
III. Dredd Blog Test Methods
The test procedures are:
1) proceed through the GBFF file searching for a CDS key that is not a 'complement', 'join', 'assembly_gap', or 'gap' (only "CDS digit..digit" references such as "CDS 15..345" are tested);
2) find the subsequent matching "/translation=" sequence of amino acids
3) proceed through the list one letter (one amino acid) at a time;
4) extract three nucleotides (one codon) from the specified DNA location for each amino acid;
5) see if that exact codon is associated with the exact specified amino acid in the Genetic Code.
If the information in the file is accurate, move on, but if there is a mismatch either in the number of CDS specifications compared to translation specifications, or in a codon to amino acid specification, add those errors to the list of errors being produced (The CDS-Translation count (column 2 in the HTML table is not a total CDS-Translation count, it is only a count of those with mismatches).
IV. Closing Comments
Here is a depiction of the CDS hypothetical example above:
...
CDS 15..345
...
/translation="MNPLAQPVIYSTIFAGTLITALSSHWFFTWVGLEMNMLAFIPVLTKKMNPRSTE
AAIKYFLTQATASMILLMAILFNNMLSGQWTMTNTTNQYSSLMIMMAMAMKLGMAP"
...
ORIGIN
AAAAATAGCCAGACATGGTGGCGGGCACCTGTAATCCCAGCTACTCGGGAAGCTGAGGC
AAGAGAATCACTTGAAACCCAGGAGGCAGAGTTTGCAGTGAGCTGAGATTATGCCACTG
CACTCCAACTTGGGCAACAGAGCGAGACTGTCTGAGAAAAAAAATACACAAAAGCACAC
ATTTTTTTATATTACAGTTGTGTAGATCAGAAGTTGGACACAGGTCTCACTGGAATAAA
ATCAAGAAGTCAGCAGGACCAGTTCCTTCTGAAGGTTTTGGGGAGAATCTGTTTCCTTA
CTGCTGCAGCTTCTAGAGGTTGCCTGTATTCCTTGGCTTGTGACCCTAATCT
TCATCTTCAAAGCCACCACTGGCAAATTTAGTCCCTTTTACGCCTGACATTT...
//
(The underlined letters (nucleotides) represent the three-letter codons in the 15..345 range).
The letters following the "/translation=" key represent amino acids (1 letter - 1 amino acid).
The Appendix files have a link to the GenBank genome sequence so that you can see the official file that was tested in each case.
If a person used a word processor to test these files it would take years to to all of them in GenBank.
The program I wrote to do this does it is seconds (less than a minute).
The next post in this series is here, the previous post in this series is here.
No comments:
Post a Comment