 |
| Fig. 1 The mRNA zone |
When samples of DNA are taken in a public place many factors challenge the ability to determine accurately what a MetaSUB techie is examining.
In fact, even in laboratory settings sometimes the reports about the nucleotides, genes, and mRNA are inaccurate because that public area is super-high intensity (see Fig. 2 for a much lower intensity example).
The issue of accuracy or the lack thereof received intense scrutiny when Sherlock Holmes type detectives started to use DNA during their detecting activities:
"It is difficult to overstate the support for DNA as it became incorporated into crime solving. Supporters of DNA for crime control celebrated its potential for the ability to control crime in a targeted and certain way. And, while DNA testing did reveal more alleged perpetrators of crime and CODIS brought standardization to the field, the many logistical and ethical problems with DNA testing quickly became clear.
These issues include basic human error and human bias, linking innocent people to crimes, privacy rights, and a surge in racial disparities.
In 2011, in their much-cited study, researchers Itiel Dror and Greg Hampikian found that DNA interpretation varied significantly among lab technicians and forensic experts. Dror and Hampikian sent the exact same DNA mixtures to 17 different experts to ascertain whether they would arrive at the same conclusion as the original forensic analysis.
Challenging the viewpoint that 'context' doesn’t matter, the 17 forensic scientists arrived at remarkably different results. Dror and Hampikian argue that this demonstrates that what the forensic scientist knows about the investigation (for example that the prosecutors are relying on the results generated to move forward) may impact the interpretation of a DNA sample. Perhaps then, it is no surprise, that there are now numerous cases of lab techs who make mistakes or argue that there was a DNA match when there was none."
(Harvard Law, 2019, emphasis added). "DNA interpretation" is a lot like Bible interpretation it would seem (Good Nomenclature: A Matter of Life and Death - 4).
This same issue was discussed the better part of a decade ago here on Dredd Blog:
"Today let's talk about a new twist, which is that there may be even more uncertainty about our genetic identity:
From biology class to “C.S.I.,” we are told again and again that our genome is at the heart of our identity. Read the sequences in the chromosomes of a single cell, and learn everything about a person’s genetic information — or, as 23andme, a prominent genetic testing company, says on its Web site, “The more you know about your DNA, the more you know about yourself.”
But scientists are discovering that — to a surprising degree — we contain genetic multitudes. Not long ago, researchers had thought it was rare for the cells in a single healthy person to differ genetically in a significant way. But scientists are finding that it’s quite common for an individual to have multiple genomes. Some people, for example, have groups of cells with mutations that are not found in the rest of the body. Some have genomes that came from other people.
Medical researchers aren’t the only scientists interested in our multitudes of personal genomes. So are forensic scientists. When they attempt to identify criminals or murder victims by matching DNA, they want to avoid being misled by the variety of genomes inside a single person.
Last year, for example, forensic scientists at the Washington State Patrol Crime Laboratory Division described how a saliva sample and a sperm sample from the same suspect in a sexual assault case didn’t match.
(The "It's In Your Genes" Myth - 2, 2013, emphasis added). This is a very important consideration when we ponder the DNA collections in public places by the MetaSUB project (those places are close to infinitely more sensitive than a table in a family's kitchen).
Hopefully we can honestly say that things have improved in the technical issues we are discussing today, however:
"But DNA technology is always advancing, and in the last decade or so, forensic experts have been using new techniques to analyze DNA mixtures, which occur when the evidence contains DNA from several people. They are also analyzing trace amounts of DNA, including the 'touch DNA' left behind when someone touches an object. These types of evidence can be far more difficult to interpret reliably than the relatively simple DNA evidence typical of earlier decades.
 |
| Fig. 2 "Location, location, location" |
With old-school DNA, the results tend to be clear cut: either a suspect’s DNA profile is found in the evidence or it isn’t, and nonexperts can readily understand what that means. [BUT] With DNA mixtures and trace DNA, the results can be ambiguous and difficult to understand, sometimes even for the experts.
(National Institute of Standards and Technology, emphasis added). That "touch DNA" evidently makes up the bulk of samples when MetaSUB folks collect it in public places.
It would seem to qualify as some "mixtures ... may be too complex to reliably interpret at all."
That will depend on the volume of the traffic at the location where the MetaSUB samples are taken, the durability of the residue under changing weather conditions, and the like.
II. Analysis Techniques
Remember the old saying "the proof of the pudding is in the eating of it", meaning that making pudding is one thing but eating the pudding determines how good it is, or if one even likes it or not.
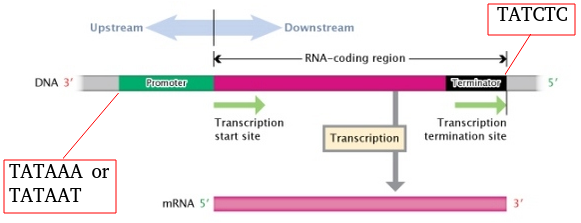
Today's appendices show clearly that the "in frame" operation depicted in Fig. 1 is a whole world of difference when compared to "out of frame" operations.
One reason is the similarity of DNA and RNA when there is no beginning or ending of a gene depicted by a promoter and a terminator, or a start or stop codon is missing.
The problem is ubiquitous:
"Structural similarities shared by the RNA polymerases (Pols) of bacteria, archaea, and eukarya reflect a deep-rooted common ancestry of transcription systems in all organisms on earth."
(Widespread Use of TATA Elements in the Core Promoters). Promoters are essential in high-quality-required circumstances:
"Core Promoter
The core promoter region is located most proximal to the start codon and contains the RNA polymerase binding site, TATA box, and transcription start site (TSS). RNA polymerase will bind to this core promoter region stably and transcription of the template strand can initiate. The TATA box is a DNA sequence (5'-TATAAA-3') within the core promoter region where general transcription factor proteins and histones can bind. Histones are proteins found in eukaryotic cells that package DNA into nucleosomes. Histone binding prevents the initiation of transcription whereas transcription factors promote the initiation of transcription. The most 3' portion (closest to the gene's start codon) of the core promoter is the TSS which is where transcription actually begins. Only eukaryotes and archaea, however, contain this TATA box. Most pAddGenerokaryotes contain a sequence thought to be functionally equivalent called the Pribnow box which usually consists of the six nucleotides, TATAAT."
(AddGene, cf. Wikipedia, TATA box). In such cases a MetaSUB techie cannot understand when an ATG codon or 3 base sequence should or should not be converted into an AUG mRna start codon (see Fig. 1).
Then again, there are various types of promoters to consider, one being the "consensus" promoter the other being the "pribnow" promoter (Wikipedia).
III. Some Analysis of MetaSUB Data
The appendices to this post show that the bulk (billions of nucleotides) have no promoter or terminator bases (Promoter-Terminator, Pribnow No Frame, Pribnow In Frame, Consensus In Frame, Codon/Nucleotide Counts, E.Coli [valid E. Coli has promoter and terminator]).
Thus, one cannot discern the place to begin to translate or transcribe the DNA into mRNA (nor the place to stop) which is a basic and necessary effort if one is to understand the transcription dynanics within microbes that are being interpreted (see the video below).
IV. Closing Comments
The MetaSUB data I have looked at so far (and have processed with genetic algorithms) indicate that it is a hodgepodge, a heap of chaotic nucleotide piles that exist in public locations.
Humans, bugs, animals have access to the public places, and so a chaotic mass of DNA and RNA molecules have been placed there.
It is not of much ultimate use at this time, IMO.
The next post in this series is here, the previous post in this series is here.